PROPHAGES OF PSEUDOMONAS AERUGINOSA
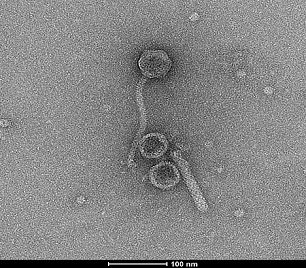
Many P. aeruginosa isolates exhibit phenotypic variation, including growth rate, virulence, motility, biofilm formation and more, while the genomic variation is considerably low. The main cause for this phenotypic variation is attributed to mobile genetic elements, particularly prophages and genomic islands. Prophages, which are bacterial viruses (bacteriophages, phages) integrated into the host's chromosome, were shown to have an essential role in horizontal gene transfer and the acquisition of new traits among the bacterial population, including metabolic and virulence factors. This project aims to examine the impact of the prophages of P. aeruginosa on the virulence and physiology of the bacteria. The focus is given on characterization of virulence factors and regulatory factors located in the prophage regions, examining the bacterial-phage relationship and understanding the prophages intra-bacterial relationship in different isolates. Characterization of prophages, especially in clinical strains of P. aeruginosa can be of service in the attempt to better understand their contribution to biofilm formation, virulence, antimicrobial resistance and more. This research may confer a new approach for dealing with infections of different P. aeruginosa strains.
